Detecting IBD2 (available from ancIBD v0.6)
Authors: Yilei Huang (yilei_huang AT eva.mpg.de)
This notebook guides you to detect IBD2 regions using ancIBD. In human genetics, IBD2 mostly occurs between full-sibling pairs. It assumes some basic familiarity with the usage of ancIBD. If you haven’t done so, we recommend you reading that first. The test data can be downloaded from this link. It contains imputed data of two pairs of full-siblings (GRG041, GRG081 and GRG007, GRG004), previously published in Rivollat et al.
Running ancIBD with its IBD2 mode
the bash command is exactly the same as for ancIBD’s default setting (aka only detecting IBD1) except for the flag –IBD2. The output will be stored in a folder called “./test” (you can change this via –out). The input files other than the vcf file of imputed genotype data can be downloaded from the link provided in the IBD1 tutorial. Change the file path below according to your own set-up.
[5]:
!for ch in {1..22}; do ancIBD-run --vcf /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz \
--out ./test --ch $ch --IBD2 \
--marker_path ./data/filters/snps_bcftools_ch$ch.csv \
--map_path ./data/v51.1_1240k.snp \
--af_path ./data/afs/v51.1_1240k_AF_ch$ch.tsv; done
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch1.1240k.vcf -T ./data/filters/snps_bcftools_ch1.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch1.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 89083 variants.
Loaded 4 individuals.
Loaded 93166 Chr.1 1240K SNPs.
Intersection 89082 out of 89083 HDF5 SNPs
Interpolating 1 variants.
Finished Chromosome 1.
Adding map to HDF5...
Intersection 88115 out of 89083 target HDF5 SNPs. 968 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch1.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch2.1240k.vcf -T ./data/filters/snps_bcftools_ch2.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch2.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 94171 variants.
Loaded 4 individuals.
Loaded 98657 Chr.2 1240K SNPs.
Intersection 94171 out of 94171 HDF5 SNPs
Finished Chromosome 2.
Adding map to HDF5...
Intersection 93469 out of 94171 target HDF5 SNPs. 702 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch2.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch3.1240k.vcf -T ./data/filters/snps_bcftools_ch3.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch3.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 77600 variants.
Loaded 4 individuals.
Loaded 81416 Chr.3 1240K SNPs.
Intersection 77600 out of 77600 HDF5 SNPs
Finished Chromosome 3.
Adding map to HDF5...
Intersection 77013 out of 77600 target HDF5 SNPs. 587 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch3.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch4.1240k.vcf -T ./data/filters/snps_bcftools_ch4.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch4.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 68720 variants.
Loaded 4 individuals.
Loaded 71634 Chr.4 1240K SNPs.
Intersection 68720 out of 68720 HDF5 SNPs
Finished Chromosome 4.
Adding map to HDF5...
Intersection 68254 out of 68720 target HDF5 SNPs. 466 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch4.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch5.1240k.vcf -T ./data/filters/snps_bcftools_ch5.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch5.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 69354 variants.
Loaded 4 individuals.
Loaded 74004 Chr.5 1240K SNPs.
Intersection 69354 out of 69354 HDF5 SNPs
Finished Chromosome 5.
Adding map to HDF5...
Intersection 68899 out of 69354 target HDF5 SNPs. 455 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch5.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch6.1240k.vcf -T ./data/filters/snps_bcftools_ch6.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch6.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 75816 variants.
Loaded 4 individuals.
Loaded 78867 Chr.6 1240K SNPs.
Intersection 75816 out of 75816 HDF5 SNPs
Finished Chromosome 6.
Adding map to HDF5...
Intersection 75059 out of 75816 target HDF5 SNPs. 757 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch6.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch7.1240k.vcf -T ./data/filters/snps_bcftools_ch7.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch7.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 59835 variants.
Loaded 4 individuals.
Loaded 62595 Chr.7 1240K SNPs.
Intersection 59835 out of 59835 HDF5 SNPs
Finished Chromosome 7.
Adding map to HDF5...
Intersection 59323 out of 59835 target HDF5 SNPs. 512 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch7.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch8.1240k.vcf -T ./data/filters/snps_bcftools_ch8.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch8.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 61089 variants.
Loaded 4 individuals.
Loaded 63916 Chr.8 1240K SNPs.
Intersection 61089 out of 61089 HDF5 SNPs
Finished Chromosome 8.
Adding map to HDF5...
Intersection 60528 out of 61089 target HDF5 SNPs. 561 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch8.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch9.1240k.vcf -T ./data/filters/snps_bcftools_ch9.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch9.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 50628 variants.
Loaded 4 individuals.
Loaded 52765 Chr.9 1240K SNPs.
Intersection 50628 out of 50628 HDF5 SNPs
Finished Chromosome 9.
Adding map to HDF5...
Intersection 50306 out of 50628 target HDF5 SNPs. 322 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch9.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch10.1240k.vcf -T ./data/filters/snps_bcftools_ch10.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch10.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 58762 variants.
Loaded 4 individuals.
Loaded 61131 Chr.10 1240K SNPs.
Intersection 58762 out of 58762 HDF5 SNPs
Finished Chromosome 10.
Adding map to HDF5...
Intersection 58363 out of 58762 target HDF5 SNPs. 399 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch10.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch11.1240k.vcf -T ./data/filters/snps_bcftools_ch11.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch11.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 54796 variants.
Loaded 4 individuals.
Loaded 57163 Chr.11 1240K SNPs.
Intersection 54796 out of 54796 HDF5 SNPs
Finished Chromosome 11.
Adding map to HDF5...
Intersection 54365 out of 54796 target HDF5 SNPs. 431 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch11.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch12.1240k.vcf -T ./data/filters/snps_bcftools_ch12.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch12.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 53884 variants.
Loaded 4 individuals.
Loaded 56133 Chr.12 1240K SNPs.
Intersection 53884 out of 53884 HDF5 SNPs
Finished Chromosome 12.
Adding map to HDF5...
Intersection 53528 out of 53884 target HDF5 SNPs. 356 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch12.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch13.1240k.vcf -T ./data/filters/snps_bcftools_ch13.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch13.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 38994 variants.
Loaded 4 individuals.
Loaded 40441 Chr.13 1240K SNPs.
Intersection 38994 out of 38994 HDF5 SNPs
Finished Chromosome 13.
Adding map to HDF5...
Intersection 38774 out of 38994 target HDF5 SNPs. 220 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch13.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch14.1240k.vcf -T ./data/filters/snps_bcftools_ch14.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch14.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 36279 variants.
Loaded 4 individuals.
Loaded 37903 Chr.14 1240K SNPs.
Intersection 36279 out of 36279 HDF5 SNPs
Finished Chromosome 14.
Adding map to HDF5...
Intersection 35743 out of 36279 target HDF5 SNPs. 536 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch14.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch15.1240k.vcf -T ./data/filters/snps_bcftools_ch15.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch15.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 34406 variants.
Loaded 4 individuals.
Loaded 35991 Chr.15 1240K SNPs.
Intersection 34406 out of 34406 HDF5 SNPs
Finished Chromosome 15.
Adding map to HDF5...
Intersection 34159 out of 34406 target HDF5 SNPs. 247 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch15.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch16.1240k.vcf -T ./data/filters/snps_bcftools_ch16.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch16.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 34373 variants.
Loaded 4 individuals.
Loaded 36000 Chr.16 1240K SNPs.
Intersection 34373 out of 34373 HDF5 SNPs
Interpolating 1 variants.
Finished Chromosome 16.
Adding map to HDF5...
Intersection 34136 out of 34373 target HDF5 SNPs. 237 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch16.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch17.1240k.vcf -T ./data/filters/snps_bcftools_ch17.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch17.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 29292 variants.
Loaded 4 individuals.
Loaded 30733 Chr.17 1240K SNPs.
Intersection 29292 out of 29292 HDF5 SNPs
Finished Chromosome 17.
Adding map to HDF5...
Intersection 28794 out of 29292 target HDF5 SNPs. 498 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch17.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch18.1240k.vcf -T ./data/filters/snps_bcftools_ch18.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch18.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 33902 variants.
Loaded 4 individuals.
Loaded 35327 Chr.18 1240K SNPs.
Intersection 33902 out of 33902 HDF5 SNPs
Finished Chromosome 18.
Adding map to HDF5...
Intersection 33720 out of 33902 target HDF5 SNPs. 182 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch18.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch19.1240k.vcf -T ./data/filters/snps_bcftools_ch19.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch19.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 18441 variants.
Loaded 4 individuals.
Loaded 19273 Chr.19 1240K SNPs.
Intersection 18441 out of 18441 HDF5 SNPs
Finished Chromosome 19.
Adding map to HDF5...
Intersection 18018 out of 18441 target HDF5 SNPs. 423 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch19.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch20.1240k.vcf -T ./data/filters/snps_bcftools_ch20.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch20.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 29052 variants.
Loaded 4 individuals.
Loaded 30377 Chr.20 1240K SNPs.
Intersection 29052 out of 29052 HDF5 SNPs
Finished Chromosome 20.
Adding map to HDF5...
Intersection 28826 out of 29052 target HDF5 SNPs. 226 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch20.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch21.1240k.vcf -T ./data/filters/snps_bcftools_ch21.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch21.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 16031 variants.
Loaded 4 individuals.
Loaded 16727 Chr.21 1240K SNPs.
Intersection 16031 out of 16031 HDF5 SNPs
Finished Chromosome 21.
Adding map to HDF5...
Intersection 15640 out of 16031 target HDF5 SNPs. 391 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch21.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Print downsampling to 1240K...
Running bash command:
bcftools view -Ov -o ./test/ch22.1240k.vcf -T ./data/filters/snps_bcftools_ch22.csv -M2 -v snps /mnt/archgen/users/yilei/Data/sex_map/prepare_IBD2_demo/gurgy.FS.vcf.gz
Finished BCF tools filtering to target markers.
Deleting previous HDF5 file at path_h5: ./test/ch22.h5...
Converting to HDF5...
Finished conversion to hdf5!
Merging in LD Map..
Lifting LD Map from eigenstrat to HDF5...
Loaded 15792 variants.
Loaded 4 individuals.
Loaded 16420 Chr.22 1240K SNPs.
Intersection 15792 out of 15792 HDF5 SNPs
Finished Chromosome 22.
Adding map to HDF5...
Intersection 15408 out of 15792 target HDF5 SNPs. 384 SNPs set to AF=0.5
Transformation complete! Find new hdf5 file at: ./test/ch22.h5
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Now let’s compute pairwise compute summary statistics. The command is the same as for the IBD1 case, except that you need the flag –IBD2 to tell the program to also summarize IBD2 sharing. The output file from this command will be stored in the current working directory unless specified otherwise via –out.
[6]:
!ancIBD-summary --tsv ./test/ch --IBD2
Chromosome 1; Loaded 17 IBD
Chromosome 2; Loaded 23 IBD
Chromosome 3; Loaded 9 IBD
Chromosome 4; Loaded 11 IBD
Chromosome 5; Loaded 22 IBD
Chromosome 6; Loaded 11 IBD
Chromosome 7; Loaded 16 IBD
Chromosome 8; Loaded 13 IBD
Chromosome 9; Loaded 6 IBD
Chromosome 10; Loaded 10 IBD
Chromosome 11; Loaded 15 IBD
Chromosome 12; Loaded 12 IBD
Chromosome 13; Loaded 12 IBD
Chromosome 14; Loaded 5 IBD
Chromosome 15; Loaded 8 IBD
Chromosome 16; Loaded 12 IBD
Chromosome 17; Loaded 10 IBD
Chromosome 18; Loaded 7 IBD
Chromosome 19; Loaded 8 IBD
Chromosome 20; Loaded 7 IBD
Chromosome 21; Loaded 3 IBD
Chromosome 22; Loaded 6 IBD
Saved 243 IBD to /mnt/archgen/users/yilei/tools/ancIBD/docs/ch_all.tsv.
/home/yilei_huang/.local/lib/python3.8/site-packages/ancIBD-0.5b0-py3.8-linux-x86_64.egg/ancIBD/IO/ind_ibd.py:25: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df["SNP_Dens"] = densities
> 8.0 cM: 149/149
Of these with suff. SNPs per cM> 220: 118/149
IBD1:
5 14
2 12
11 10
7 9
3 7
13 7
10 7
1 6
4 6
6 6
16 6
12 5
9 5
15 3
8 3
18 3
14 2
17 2
20 2
19 1
21 1
22 1
Name: ch, dtype: int64
IBD2:
8 10
2 9
1 7
7 7
17 6
11 5
20 5
13 5
12 5
6 5
5 4
16 4
22 4
10 3
15 3
4 3
14 2
18 2
19 2
9 1
3 1
21 1
Name: ch, dtype: int64
iid1 iid2 lengthM
0 GRG007 GRG004 7.356109
1 GRG041 GRG081 8.424912
[735.610883682966, 0.0, 0.0, 842.4912158669028, 0.0, 0.0]
Saved 6 individual IBD pairs to: /mnt/archgen/users/yilei/tools/ancIBD/docs/ibd_ind.tsv
Visualize IBD2 region with imputed data
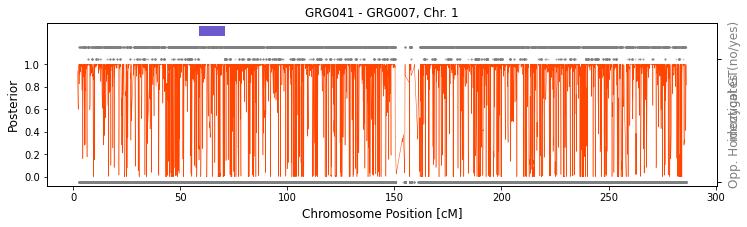
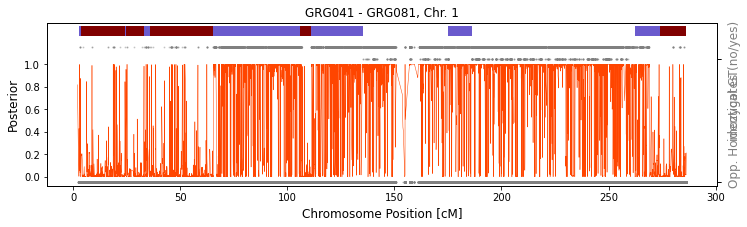
In some cases it might be beneficial to visually examine the detected IBD2 region to make sure it indeed looks like real IBD2. We can use the function ancIBD.run.run_plot_pair_IBD2 to do just that. The function will call IBD2 on a given chromosome and visualize the called IBD1 as blue bar on top of the figure and called IBD2 as dark brown bar (which is then plotted on top of the IBD1 bar). There are two rows of gray dots below the IBD bar. The higher row depicts sites where the two
individuals have different imputed genotypes (thus cannot be IBD2). The lower row depicts sites where the two individuals have oposing homozygotes (thus cannot be IBD1).
[3]:
from ancIBD.run import run_plot_pair_IBD2
Let’s first visualize a full-sib pair, using chr1 as an example.
[2]:
run_plot_pair_IBD2(path_h5="./test/ch",
iids = ["GRG041", "GRG081"], ch=1,
plot=True)
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Filtering to 0.99 GP variants: 0.931x
Filtering to 0.99 GP variants: 0.869x
Filtering to common GP variants: 0.836x
Filtering to 0.99 GP variants: 0.931x
Filtering to 0.99 GP variants: 0.869x
Filtering to common GP variants: 0.836x
Now let’s visualzie a non full-sib pair. We can clearly see the absence of IBD2 by looking at the higher row of gray dots, which is densely distributed across the entire chromosome.
[4]:
run_plot_pair_IBD2(path_h5="./test/ch",
iids = ["GRG041", "GRG007"], ch=1,
plot=True)
Attention: Some data in GP field is missing. Ideally, all GP entries are set.
Filtering to 0.99 GP variants: 0.931x
Filtering to 0.99 GP variants: 0.705x
Filtering to common GP variants: 0.681x
Filtering to 0.99 GP variants: 0.931x
Filtering to 0.99 GP variants: 0.705x
Filtering to common GP variants: 0.681x